|
|
|
|
|
|
 |
 |
|
|
|
|
|
|
 |
 |
The terminating step of the divide and conquer algorithm is:
a) that point at which protein synthesis is
disabled by the technique. OR
b) the point at which the Normal DNA and Tumor DNA
are determined to be identical.
Every cell requires growth signaling. Broken growth signaling may occur, not as the Boolean of an OFF or ON, but rather as the Continuous or Intermittent Over or Under expression of a signaling substance. If this is the case then n, the number of proteins made by normal DNA will equal m, the number of proteins made by tumor DNA and a Boolean approach will fail. Further the entries in each list will be identical. If unique proteins arise, such as in myeloma this is ideal since we would like to associate particular substances with the pathological state.
| Protein Arity: Three Possible Outcomes | ||||||||||||
|
| Example: Same Number Does Not Mean Same Proteins | ||||
Protein count is the same, protein type is not. |
It was suggested to Munshi that PCR/cDNA amplification could be used to recover the genetic machinery that gave rise to the proteins. This would salvage a particular sample I had hoped to tissue culture but was damaged by freezing.
Note that this work does not attempt to map parts of the normal tissue gene that may carry defects in tumor suppressor genes such as BRCA1. Rather the purpose here is to compare tumor DNA and normal DNA with the sole intention of differentiating tumor protein synthesis and expression from normal tissue protein synthesis and expression.
Microdissection:
It was implied to me by Munshi that microdissection of the sample would
decrease the likelihood of cross contamination. UAMS has recently
acquired a confocal microscope that could be helpful to this end.
For example. If normal tissue DNA was found to be mixed with the
tumor tissue DNA, then both types of DNA would be amplified. Contrawise
if tumor tissue DNA contaminated the normal tissue DNA then again both
types of DNA would be amplified. I will prove below that one sided
contamination from either side is allowable, but two sided cross contamination
is not.
Contamination:
There are three contamination cases. The term one-sided contamination
indicates that A has traces of B, but B does not have any trace of A.
The same remark applies if we switch the roles of A and B. First a quick
review of Boolean algebra.
Boolean Algebra:
There are three fundamental operators in Boolean algebra, AND, OR &
NOT:
|
Operation
|
|
Symbol |
|
|
| set intersection |
|
|
|
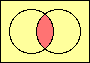 |
|
set union
|
|
|
|
 |
|
set negation
|
|
|
|
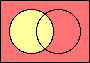 |
|
set difference
|
MINUS |
|
|
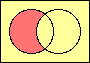 |
Definitions:
A = List of Proteins Generated by Normal DNA
B = List of Proteins Generated by Tumor DNA
A' = (A | B) = List of Proteins Generated by Normal DNA contaminated
with Tumor DNA)
Case 1: We will begin with the least likely case, one-sided contamination of A with B. Imagine that due to specimen contamination, we find ourselves in Case 3 of the protein arity table above. List A is contaminated to include List B as a subset. We call this contaminated list List A', the prime being the contamination operator. We seek to subtract List B from List A' to obtain a list of proteins that are only in the clear margin, i.e. the true List A. Or more succinctly:
|
|
Now having recovered the true A, we can compare this list with the true B and proceed.
Special Circumstances:
Two circumstances could bring us into the A = {a, b, c, d, e, f} and
B = {d, e, f} case.
1) A was contaminated with B, meaning A is really A' OR
2) A, the Normal tissue makes three more proteins than B, tumor tissue.
We can't always determine which of the two special circumstances is
in force. This will be termed aliasing.
Note also that we are now tacitly equating a tissue type with the
proteins it produces, which if you think about it seems reasonable.
Case 2:
Imagine that we switch the roles of A and B, so that A is tumor protein
list and B is normal protein list. We can now run through the same
argument again. This is the second case, one sided contamination pointing
the other direction. We get this work for free by switching the roles
of normal tissue and tumor tissue. This is the more likely case,
where we got clear surrounding margin tissue, but when sampling the tumor,
some normal tissue was inadvertently included.
Case 3:
In this case both A and B are contaminated with each other to produce the lists
A' and B'. Now we presumably cannot recover any proteins unique to A and
B because the Boolean rules that governed the previous situation now have to
be promoted to continuum rules where we talk about amounts of different proteins
and begin to use words like under and overexpression of proteins. Differential
amplification (making more tissue specific protein if more of a given tissue
type is present in a mixed sample.) would also be useful if our Boolean thesis
failed and we were in a pure under or overexpression situation, without the
clear absence or presence of tumor specific proteins.
There are three difficult problems that must be solved for Lynn to be healed of breast cancer in the most permanent sense:
It is also helpful to understand currently mapped landmarks of genetic origin that may apply to Lynn's specific cancer. These include BRACA1 & 2 (for which she has not been tested) and Her2Nu (for which she has been tested and is positive). I must confess at this writing my understanding on whether Her2Nu is a receptor issue or a genetic defect issue is lacking. It is helpful not only to understand these landmarks, but the process that gave rise to their discovery.
Sometimes it is helpful to consider unifying apparently different disciplines, for example the mapping of combinatorial problems onto genetic sequences could be combined with PCR amplification in a macro level process of resolve and amplify, resolve and amplify. In this case we can begin to think of PCR and combinatorial mapping as computational operators. Process design can then be thought of as mathematical equation construction, rearrangement, solution and optimization. We can work in symbol space when that is to our advantage, in numerical space and in reagent space in a manner that maximizes the contribution of each.
A helpful exercise along this line, is to consider classic sorting and search problems, embedded as sequence amplification, combination and rearrangement.