Fall
1998
Genetic Biochemistry
- Lecture 1
Intracellular compartments
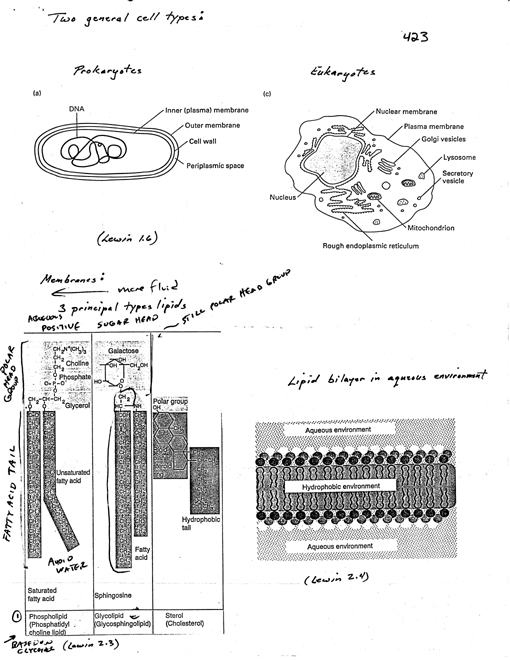
Two general cell types [1 Lewin 1.6]
Prokaryotes (bacteria)
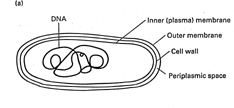
· normally
single cell compartment.,
· bounded
by membrane(s) - give security against outside world
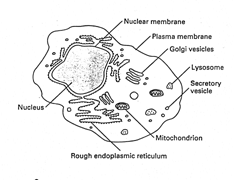 Eukaryotes
Eukaryotes
· division
of each cell into nucleus (contains genetic material) surrounded by cyto,
which is bounded by PM.
· cyto.
contains other discrete compartments bounded by membranes
· Membranes
· characteristic
properties result from high lipid content · crucial feature
of lipids - amphipathic
· one
end - polar "head"
· other
end - hydrophobic "tail(s)"
· major
bulk of lipid
· differ
over all length, nature of C-C bond (sat'd. or unsaturated)
· rotation
restricted wi C=C, has a bend
· three
principal types of lipids [1 Lewin, 2.3]
· (1)
phospholipids
· head
has pos. charge group linked via neg. charge phosphate to rest
molecule
· e.g.,
PC - choline-phosphate glycerol affached to 2 hydrophobic tails
· lipids
based on glycerol have one satd., one unsatd fa tail ·
(2) glycolipids
· presence
of oligosaccharide
· chain
of sugars typically 1-15 residues
· in
animal cells, connection biw sugar and f.a. tail is sphingosine
(long amino alc.)
· lipids based
on sphingosine have f.a. chain in addition to f.a. of sphingosine
· in plants and bacteria, glycerol connects head and
tail
· (3) sterols
· contain steroid
ring -
1
· give rigidity because
steroid ring is planar
· cholesterol, prominent
in animal cell membranes, has polar -OH group at term.
· in aqueous environ.,
lipid has polar head exposed, tries to bury hydrophobic tail
· lipid bilayer [1 Lewin
2.4]
· in diff. membranes,
lipid composition varies considerably
· both ratio of protein
to lipid
· types of lipid
· tf; diff. membranes
have diff. biophysical properties
· imp. property of membrane,
lipids can move laterally (not between layers) - fluidity
· more readily tails
of adj. lipids pack, more crystalline structure (less fluid) ?
· depends on length
and type of lipid tails which more f lui SD -
· unsaturated chains
more difficult to pack, more fluid
· PM of animal cells
relatively large anits cholesterol - increases
mechanical stability
· plant cells lack
cholesterol, but have other sterols
· PM only membrane
to contain signif. amt glycolipids
· consider membranes
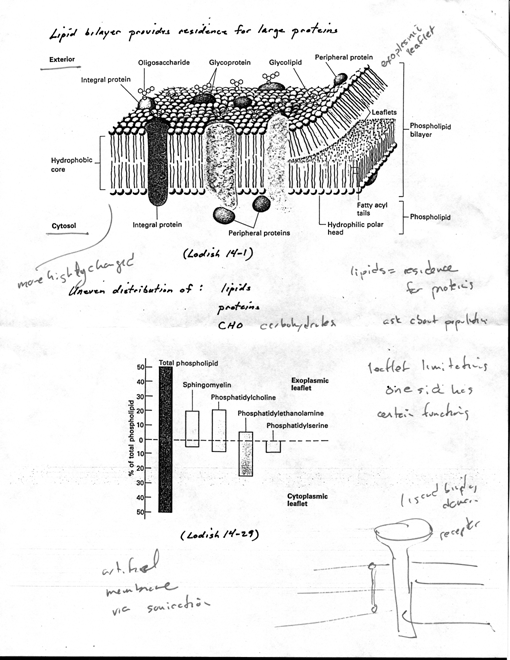
as lipid bilayer to provide residence for large protein
molecules [2 Lodish 14-1]
· proteins can move
laterally, but difflise more slowly than lipid
· proteins can be
internalized due to stimulus, others pass thru
membrane as secreted, tf; proteins
associated with bilayer dynamic
· transmembrane protein
(e.g., receptors)
· membrane has two "faces"
· cytosolic face
· noncytosolic (called
diff. names depending where it is)
· PM, extracellular
environment
· organelle, lumen
· uneven distribution of
three components [2 Lodish 14-29]
· (1) diff. lipids in
diff. monolayers (1,oth polar head group and tails)
· lipids on cytoplasmic
face more highly charged, tend to be unsat
(more 2
· (2) proteins
oriented so diff. sequences present on each face
· (3) CHO groups
(glycolipids or glycoproteins) found exclusively on extracellular
face
· PM, exterior of cell rich
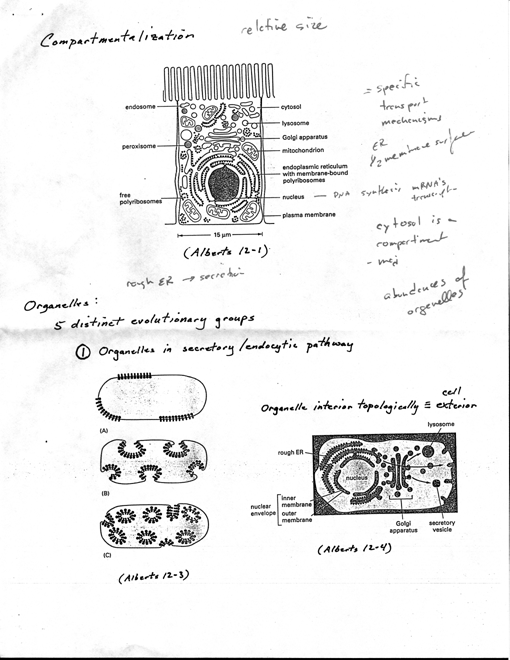
in oligosaccharides (cell adhesion, cell-cell recog.) · Compartmentalization
[3 Alberts 12-1]
· eukaryotic cells
subdivided into flinctionally distinct, membrane-bound compartments
· lipid bilayer impermeable to most hydrophilic molecules,
tf, each must contain
transport proteins for import/export
of specific metabolites
· must have mechanism
to import & incorporate specific proteins that make organelle
unique
· each has own characteristic
set of enzymes, other specialized molecules
· distribution systems
transport specific products from one compartment to another
· Major compartments
- all eukaryotic cells have same basic set · Nucleus
· major site of DNA
and RNA synthesis · Cytoplasm
· little more than
half total volume of cell
· site of protein
synthesis
· most of intermediary
metabolism (small molecule degradation and synthesis
to provide building blocks
of macromolecules)
· ER
· about half total
area of membrane
· membrane bound ribosomes,
synthesis of integral membrane proteins and
soluble proteins (most destined
for secretion or other organelles)
· this is important
difference biw how proteins directed to ER and other
cytoplasmic organelles
· proteins translocated
to ER during synthesis, to other organelles only
after synthesis complete
· produces lipid for rest
of cell
· flinctions as
store for Ca2+
- ·- Golgi apparatus
3
· stacks of
cisternae
· receives
lipids and proteins from ER, dispatches to other destinations
(usually wi covalent modifications
· mitochondria (chloroplasts
in plants)
· generate most of
ATP to drive cellular rxns · lysosomes
· digestive
enzymes to degrade defiinct intracellular organelles, macromolecules
and particulates endocytosed
· endosomes
· intermediates during
endocytosis · peroxisomes
· enzymes
utilized in oxidative rxns
· Although each organelle
carries out same basic flinctions in all cell types, can vary in abundance
and have additional properties in different cell types. (secretory cell
1
more RER · Topological Relationships
· Precursors to first
euk. cells thought simple organisms like bacteria (1 2- 3) ·
plasma membrane, no internal membranes
· PM provide all membrane-dependent
flinctions
· pumping
ions, ATP synthesis, protein secretion, lipid synthesis
· proflision of internal
membrane adaptation to increase in size (1K-i OK x larger volume than
bacteria, 10-30 x larger in linear dimension), tf; smaller ratio surface
to volume and PM too small for the many vital functions.
O Area = 1 fi Area= 10 4pir^2
Volume=.093 ½½ Volume=2.97 4/3pir^3
A/V=10.7 A/V=3.4
If cell area is lOx larger, the volume
is 32x larger (A/V 10.7 ® 3.4).
4
· 5 distinct evolutionary
groups
· (1) All organelles
that flinction in secretory and endocytic pathway (ER, Golgi, endo. lysosomes,
transport vesicles
· [3 Alberts 12-3],
bacteria with specialized membrane patches ("purple membrane" containing
bacteriorhodopsin in Halobacterium), represent primitive organelles.
In some photosynthetic bacteria invaginated PM, others invaginations
seems pinched off completely forming sealed membrane-bound vesicles
specialized for photosynthesis
· If euk. organelle
originated by this pathway, interior topologically equivalent to exterior
of cell. [3 Alberts 12-4]
· interiors communicate
extensively with one another and with outside of cell · (2)
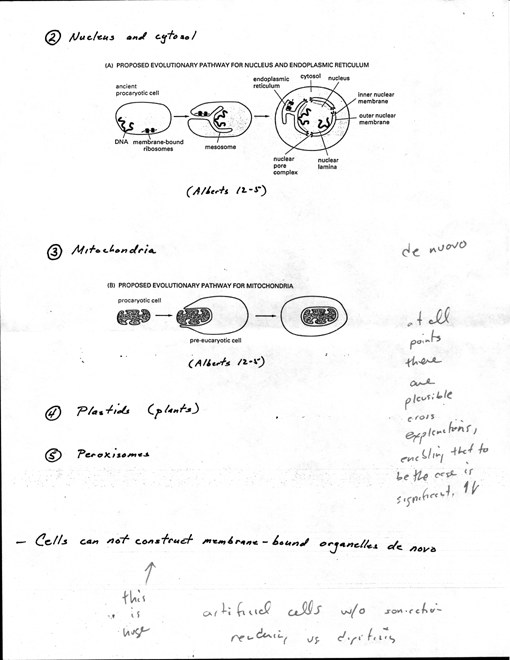
Nucleus and cytosol
· Evolutionary
scheme reasonable explanation for cell nucleus with double membrane.
[4 Alberts 12-5A]. Bacteria, single chrom. attached to special sites
inside PM. possible that double4ayered nuclear envelope originated
as deep invagination of PM.
· ribosomes attached
to cytosolic face of PM in bacteria, evolutionary origin of ER membrane
from PM may explain why ribosomes attached to ER of euk. cells
· Also, explains
why nuclear compartment topologically equiv. to cytosol. (1n higher
euk, during mitosis nuclear envelope breaks down releasing contents
into cytosol - not seen for other membrane-bound organelles).
· Space blw two
nuclear membranes topologically equiv to exterior of cell and is continuous
with lumen of ER
· (3) Mitochondria
· contain own genomes
(different than other membrane-bound organelles) · Nature
of genomes and resemblance of proteins to some present-day bacteria
suggest mito (& plastids)
evolved from bacteria engulfed by other cells [4 Alberts 12-SB]
· Inner membrane
of mito corresponds to original PM of bacterium, lumen evolved from
bacterial cytosol
5
· May explain
why lumen of mito. remain isolated from vesicular traffic that
connects lumens of other compartments and outside of cell.
· (4) Plastids
- plants only · like mitochondria
· (5) Peroxisomes
· Cells can not construct
membrane-bound organelles de novo · when cell divides,
must duplicate membrane-bound organelles
· enlarge existing
organelles by incorporation of new components
· organelles then
divide and distribute to two daughter cells, thus each daughter cell
inherits complete set of specialized membranes
· information required
to construct membrane-bounded organelle does not reside exclusively in
the DNA that specifies organelle's proteins (DNA can direct expression
of protein with targeting information, but without signal receptor it
would have no place to go)
· Must have info in
form of at least one distinct protein that preexists in organelle membrane,
and this info passed from parent to progeny in form of organelle itself
· Essential for propagation
of cell's compartmental organization (just like DNA essential for propagation
of nucleotide and aa seq)
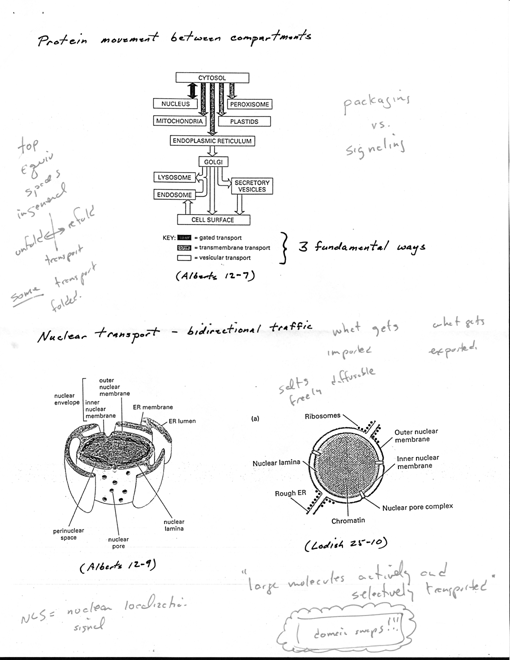
· Protein movement between
compartments [5 Alberts 12-7]
· all proteins are
synthesized on ribosomes in cytosol except few syn. on ribosomes of mito
and plastids
· fate depends on
sorting signals - direct delivery to locations outside of cytosol
· most proteins
do not have sorting signal, remain in cytosol
· others have specific
signals for nucleus, ER mito, plastids, peroxisomes
· can also sort
from ER to other destinations in cell
· 3 fundamental ways for
movement from one compartment to another
· (1) gated transport
- protein traffic biw cytosol and nucleus (topo equiv spaces) in
continuity through nuclear
pore complexes [3 Alberts 12-4]
· (2) transmembrane
transport - membrane-bound protein translocators directly
transport specific proteins
across a membrane from cytosol to space topo distinct
· transported
protein must unfold to go through membrane
· initial transport
of proteins from cytosol into ER lumen or mito
6
· (3) vesicular
transport - vesicles carry proteins from one compartment to another
· e.g., transport from ER to Golgi apparatus
· transported
proteins do not cross a membrane, move only biw topo equiv compartments
· Summary [5 Alberts
12-7]
· each mode usually
selective by sorting signals in transported protein, recognized by
complementary receptor proteins in target organelle
· Nuclear transport [5
Alberts 12-7]
· nuclear envelope
formed from two concentric membranes that are continuous w/ER
· two membranes maintain
distinct protein composition [5 Alberts 12-9, Lodish 25-10]
· inner nuclear membrane
· contains specific proteins
act as binding sites for nuclear lamina that supports it · outer
nuclear membrane
· closely resembles
membrane of rough ER
· like RER, outer
nuclear membrane studded with ribosomes engaged in protein synthesis
· these proteins
transported into perinuclear space, which is continuous with ER
lumen
· bidirectional traffics
occurs continuously biw cyto and nucleus
· many proteins
that fn in nucleus (histones, DNA/RNA polymerases, transcription factors,
RNA-processing proteins) syn. in cytosol, selectively imported into
nuclear compartment
· at same time,
tRNAs and "iRNAs syn in nuclear compartment exported to cytosol
· export also selective, niRNAs only exported if properly
processed
· sometimes complex
transport
· ribosomal
proteins syn in cyto, transport to nucleus, assemble with rRNA
into particles, then exported again into cyto as ribosomal subunit
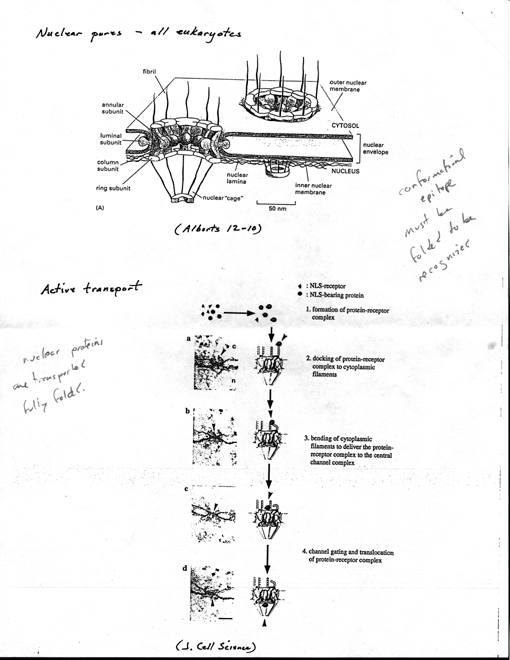
· Nuclear pores - all
eukaryotes
· Formed by large structure
known as nuclear pore complex [6 Alberts 12-10] · each pore
contains one or more aqueous channels for passive diffusion of water-
soluble molecules
7
· Many proteins too
large to pass by diffusion, so envelope allows different protein composition
(ba r ri er - gate keeper
· proteins destined
for nucleus, tf, must posses nuclear localization signals that bind
to specific receptor proteins in pore complexes and are actively transported
across nuclear envelope
· nuclear localization signals
(NLS)
· when nuclear proteins
extracted from nucleus and microinjected back into cyto, even large ones
efficiently reaccumulate in nucleus
· selectivity of nuclear
protein import resides in nuclear localization signals
(present only in nuclear proteins)
· defined by recombinant
DNA technology - domain swap (explain)
· normally accumulates
in nucleus shortly after cyto syn
· single aa mutation
prevented nuclear import
· used short lengths
of DNA encoding region around mutation to define
localization signal fused to
cytosolic protein
· Active transport
· visualized by coating
gold particles wi nuclear proteins, injecting into cyto, follow fate by
EM [6 JCS]
· initial interaction
reqs one or more cytosolic proteins that bind to NLS, help
direct nuclear protein to pore
complex (appears to bind to fibrils that project
from rim of complex)
· nuclear protein
moves to center of pore complex, actively transported by
process that requires ATP
· using various
sized gold beads, opening can dilate ii~
·
· Export of new ribosomal
subunits and mRNA also depends on selective transport
· 20-nm dia gold
spheres coated wi small RNA molecules (5S or tRNA),
injected into nucleus (frog
oocyte), rapidly transported through nuclear pores
into cyto
· if injected into
cyto, remain there; tf; seems pore contains receptors that
recognize RNA molecules (or
proteins bound to them) destined for cyto
8
· use different
size gold particles, one coat wi RNA inject into nucleus, other coat
winuclear import signals inject cyto; show that single pore complex
allows traffic both directions
· mechanism fundamentally
different from transport across membranes of other organelles - occurs
through large, regulated aqueous pores rather than through a protein
transporter that spans one or more lipid bilayer
· nuclear
protein transported fully folded (e.g., newly formed ribosomal
subunits transported as assembled particle)
· other organelles,
proteins unfolded during transport (maybe mito exception)
· ER
· plays central part
in lipid and protein biosynthesis
· its membrane is site
of pdtion of all transmembrane proteins and lipids for most organelles
· also makes major contribution
to mito and perox membranes by pdcing most of their lipids
· ER captures selected
proteins from cyto as they are being synthesized
· two types of proteins
[7 Lodish 16-1]
· transmembrane
proteins, only partly translocated across ER membrane, become
embedded in it
· some will remain
in ER, many destined to reside in PM or membrane of
another organelle
· water soluble
proteins, fully translocated, released into lumen of ER
· destined for
lumen of organelle or for secretion
· all these proteins, regardless
of fate, directed to ER membrane by same kind of signal peptide
and translocated by same mechanism
· import begins
before polypeptide chain completely syn - co-translationally
· different than
import into mito, chloroplasts, nuclei, peroxisomes - posttranslational
and requires different signal
· never released
into cytosol; tf, never folds before reaching translocator in membrane
· in contrast, posttranslational
import into mito, chioro, cytosolic chaperones required
to keep unfolded
· ribosome synthesizing
protein directly attached to ER membrane - create regions
termed RER
9
· two spatially separate
populations of ribosomes [7 Alberts 12-33]
· membrane-bound, cytosolic
side of ER membrane
· free ribosomes, unattached
to any membrane
· differ only in proteins
that they are making at a given time
· when rib
making protein wIER signal peptide, signal directs rib to ER membrane
· many rib
can bind single mRNA, polyrib usually formed attached to ER membrane
· individual
ribs return to cyto when finished translation, mRNA tends to remain
attached to ER by changing population of ribs
· polyribs
also form wi mRNAs lack ER signal, remain free in cyto, protein
pdc discharge in cyto
· signalhypothesis
· signal hypothesis
- leader serves as signal peptide to direct secreted protein to ER membrane,
cleaved off by signal peptidase in ER membrane before polypeptide chain
completed [8 Lodish 16-11]] (un like NLS)
· ER signal peptide
guided to ER membrane by at least two components · signal-recognition
particle (SRP)
· cycles biw ER membrane
and cytosol, binds to signal peptide · SRP receptor (aka docking
protein)
· SRP ribosome
complex binds to SRP receptor, integral membrane protein (2
subunits) exposed on cyto surface
of RER
· Sequence of events
· signal seq cleaved
off in lumen by signal peptidase, quickly degraded
· peptide chain continues
elongate, extruded through ER mem
· Bip binds to exposed
hydrophobic segments
· prevent denaturation
or aggregation
· ATP hydrolysis drives
Bip release
·
OK
propcrly, not
10
· ~~aiibe of to
· small minority of proteins
shown to be imported into ER posttranslationally · e.g., yeast
alpha mating factor
· as with mito,
require cyto chaperones to prevent folding
· like mito and chloroplast,
signal peptides cleaved by signal peptidase on luminal side of ER membrane
· signal peptide,
by itself, not sufficient to signal cleavage by peptidase; requires
adjacent cleavage site recognized by peptidase
· internal ER
signal peptides do not contain these additional sites, tf, not cleaved
- serve to retain transmembrane protein in lipid bilayer
· single-pass transmembrane
protein [8 Lodish 16-17] · Multipass transmembrane proteins
· polypeptide chain passes
back and forth repeatedly across lipid bilayer [8 AAlberts 12-
47]
· thought that internal
signal peptide serves as start-transfer signal to initiate translocation,
this continues until stop-transfer peptide reached
· Post-translational modification
of secretory and membrane proteins in RER · Five principal modifications
during transit to cell surface
· (1) formation of disulfide
bonds - ER
· formation of disulfide
bonds in lumen of RER, never cytosol
· confined to
secretory proteins, luminal/extracellular domains of membrane proteins
· GSH major thiol
containing molecule in euk cells [9 Lodish 16-22] ·
catalyze formation in ER
· two
thiol-disulfide exchange rxns
· lumen
of ER, GSH:GSSG ratio ~5:1, optimal for formation of disulfide
bonds
· prevent
formation of disulfides in cytosol · [GSH] in
cytosol ~1 0mM
· GSH:GSSG
ratio ~ 50:1, drive rxii to left (toward Cys-SH,
away from Cys-S-S-Cys)
11
· almost all
cysteine residues in protein domains exposed to either extracellular
space or lumen of organelles in exo or endo pathway are disulfide
bonded
· disulfide
bonds do not form in domains exposed to cytosol due to reducing
environment
· bacteria
reducing environment, can't use for synthesis of mammalian proteins
normally stabilized by disulfide bonds
· (2) proper folding
of polypeptide - ER · Bip (binding protein)
· peptidyl-prolyl isomerase
- catalyze rotation of exposed pep-pro bond · (3) formation
of multichain proteins - ER
· e.g., assembly of Ab,
hemagluttin (HA) precursor [9 Lodish 16-24] · (4) addition and
modification of carbohydrates
· Most proteins
synthesized in RER are glycosylated by addition of common N4inked
oligosaccharide
· glycosylation
is one of major biosynthetic fns of ER
· most soluble
and membrane-bound proteins syn in ER are glycosylated
· few proteins
in cytosol glycosylated (those that are have simpler modification
-- N-acetylglucosamine of Ser or Thr)
· (5) specific proteolytic cleavages
· transport from ER to
Golgi referred to default pathway [10 AAlberts 13-3]
· proteins do not
seem to require specific signals - any protein that enters ER (folds and
assembles properly), automatically transport through Golgi to cell surface
unless signals detain in earlier compartment or divert it
· quality checkpoint
for proper folding and assembly (don't want misfolded proteins to
reach cell surface, may be
recognize as foreign, might stimulate immunologic
response
· tf, ER one of maj
or sites where proteins degraded
· Golgi apparatus [10
Alberts 13-4]
· two distinct
faces (cis - entry face, trans - exit face)
· both closely
connected to special compartments - network of interconnected
tubularstructures
12
· cis GoAlgi
netvork (intermediary or salvage compartment)
· trans GoAlgi
network
· both thought
important for sorting (e.g, proteins entering CGN can either move
onward or return to ER; proteins exit TGN sorted by destination -Alysosomes,
secretory vesicles, cell surface)
· cis compartment thought
continuous wi CGN
· next compartment,
medial - central cisternae of stack
· trans compartment,
final site of glycosylation - lumen thought continuous w/TGN · Oligosaccharide
processing in Golgi
· two broad classes
of N-linked oligo [11 Lodish 16-27]
· high-mannose
oAligosaccharides
· no new sugars
added in Golgi
· contain 2 GlcNAc,
many mannose residues
· compAlex oAligosaccharides
· more than original
2 GlcNAc, variable number galactose, sialic acid
(only sugar in glycoproteins
winet negative charge), and sometimes
fucose
· formed by combination
of trimming of original oligo and addition of
other sugars
· which processing determined
mainly by configuration of protein
· if oligo on protein
accessible to enzymes in Golgi, then likely converted to
complex form
· protein may
fold rapidly, so site not accessible
· if inaccessible,
likely remain high-mannose form
· each cell type
has its own set processing enzymes, thus same protein may be
modified differently in different
cells
· processing follows
highly ordered pathway [11 AAlberts 13-11]
· each cisterna
contain own set of processing enzymes
· proteins modified
in successive stages as move from one cisterna to next
· only accepted
as substrate if properly processed by preceding enzyme
· forward movement mediate
by transport vesicles
13
· like
ER to Golgi, thought nonselective · functional compartrnentalization
· sensitivity/resistance
to specific glycosidases used as marker for modifications ·
not only N-linked oligos altered in Golgi, many other modifications
· sugars
added to OH groups of selected Ser/Thr residues
· 0-linked
glycosyAlation - series of glycosyl transferases using sugar
nucleotides in lumen of Golgi
· some
Tyr residues sulfated in lumen of TGN · Asymmetrical distribution
· oligo
chains added to luminal side, tf, distribution of carbohydrate on
membrane proteins and lipids asymmetrical
· topology
maintained during transport ~odish 16-1]
· oligo
of 41 glycoproteins and glycolipids in intracellular membranes
face lumen, those in PM face outside
· Vesicular
Transport
· transport
vesicles bud off one compartment fuse with another [12 AAlberts 13-2]
· most
transport vesicles form from specialized coated regions of membranes
· before
vesicle can fuse with target membrane, coat discarded so two membranes
interact directly
·
two well-characterized types coated vesicles [12 AAlberts 13-3]
· cAlathrin-coated
vesicAles
· mediate
selective transport of transmembrane receptors (M6P receptor
from TGN to lysosome or LDL receptor from PM)
· coatomer-coated
vesicAles
·
mediate nonselective transport of default pathway ·
maybe third type: calveolin-coated vesicles
· PM
most cells has morphologically, biochemically distinct invaginations
- caveolae
· fn
uncertain, may bud off to form vesicles
· Unidirectional
vesicular transport requires chemical energy
14
· concentrating
proteins against gradient
· otherwise,
proteins would reach equilibrium b/w compartments · transport
from TGN to cell surface
· vesicles
destined for PM normally leave TGN in steady stream
· membrane
proteins and lipids provide new components for cell PM
· soluble
proteins secreted to extracellular space
· fusion of
vesicles w/PM called exocytosis
· all
cells require constitutive secretory pathway (13)
· secondary
pathway in specialized secretory cells, soluble proteins and other
substances sorted
into secretory vesicles for later release - reguAlated secretory pathway
· hormones,
neurotransmitters, digestive enzymes · Endocytosis
· two main
types, based on size of endo vesicle
· phagocytosis
- ingestion of large particles
· large vesicles
(phagosomes), general> 250 nm
· imp in animal
cells for purposes other than nutrition
· normally specialized
phagocytic cells
· mammals, two
classes wbc "professional" phagocytes
· macrophage -
widely distributed in tissues and blood
· neutrophils
· defend against
infection, digest invading microorganisms
· macros also
scavenge senescent, damaged cells, debris
· quantitatively
more imp
· phagocytose>
1011 senescent rbc each day
· pinocytosis - ingestion
of fluid and solutes
· small vesicles
(<=150 nm)
· extracellular
fluid trapped, substances dissolved in fluid internalized - called
fluid-phase endocytosis
· most of
their PM
15
· by cell
· e g , ingest
25% own volume of fluid/hour
· 3% Iniri, or 100%
~ 30
· cell surface area and
volume remain constant, linked to exo.
· endosomaAl compartment
- complex set of heterogeneous membrane-bound tubes
and vesicles, extend from periphery
to perinuclear region (12 Alberts 13 -3)
· often close to Golgi,
but distinct
· two set of endosomes
readily distinguished
· earAly endosomes -just
beneath PM
· tracers appear within
a minute or so
· Alate endosomes - close
to Golgi and near nucleus
· tracer appears after
5-15 min
· interior of
endo kept acidic (pH 6) by driven H+
· than
· early endo acts as main
sorting station (like TGN in exo pathway)
· many ligands released
in acidic environment
· ligands released
in early endo usually degraded in lysosomes
· other ligands remain
bound, share fate wireceptor
· (1) most receptors
return to PM domain they came from
· LDL receptor
· receptor recycled
to PM
· LDL goes to
lysosome
· transferrin
receptor pathway similar, except transferrin
stays bound to receptor, releases
iron in endosome,
returns to PM, released in
neutral pH of extracellular
fluid
· (2) some go to
lysosome for degradation
· EGF receptor
clusters in pits only after binding ligand
· most do not
recycle, degraded in lysosome
16
· EGF binding, tf;
leads to decrease in conc. of receptors on cell surface - receptor downregulation
· (3) some go to different
domain of PM - transcytosis
17
Directed Searches:
Golgi
vesicles: Think of Golgi
bodies as protein post offices preparing proteins for transport.
Nuclear Membrane
Lysosome:
The cellular composter, breaks
down waste materials to their components, pH=5.
Secretory
vesicle: Organelles responsible for hormone et. al. transport via exocytosis.
Ref
A. Ref B.
Mitochondrion:
This double membrane organelles are the
batteries of the cell. Convert glucose and oxygen to ATP.
Krebs cycle and glycolysis pathways
are active within mitochondria.
Endoplasmic
Reticulum: A three
dimensional maze where:
a) large molecules are transported
b) proteins are stored
c) ribosomes are attached
d) fatty acids and lipids are synthesized.

 Eukaryotes
Eukaryotes




