| Cell
World Storyboard: The
DNA Roller Coaster
Characters:
RNA Polymerase, Mattie, the gang Segment
Sound Track: "Save
Yourself" Car
Detail: RNA Polymerase | |
Action:
The roller coaster breaks at the line: "I cannot save
you, I can't even save myself" |
|
 |
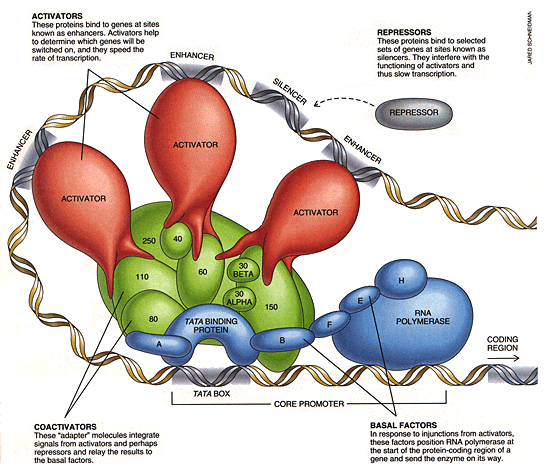 Image Source: Robert Tjian: Scientific American Feb 1995 |
Backstory - A Conversation With Barry Hurlburt on DNA Traffic Jams We
slip back into the science talking about the way that
proteins slide along DNA. I always thought of DNA as “referenced
when needed”. Barry describes it as, "A Hong Kong street
so crowded with proteins that you cannot even see the road
anymore", the diametric opposite of “Grand Central
Station empty” my favorite Life magazine
image. Barry points out
that if you time-lapse it, “everybody gets where they’re ‘sposed
to” and I infer, “maybe you can even see the street
a little”. The intracellular gook is so thick, so viscous
you would have a hard time walking through it. So, one thing leads to another, and I love questioning smart people until they say, “It isn’t known” because that’s what getting a Ph.D’s is ‘sposed to be about right? Anyhow. I ask Barry, “If DNA is COVERED with these protein pachyderms’, what happens when one protein runs into another and wants to go on, but it can’t because somebody is stalled on the track up ahead? Automaton view: A strand of DNA is essentially a sequential access (not random access) medium. It is a Turing tape, a consecutive set of instructions that must obey certain rules. (Turing tapes can back up, I wonder if the DNA reader can?) And in the space of rules, sequentiality puts serious constraints on what can happen. Here’s what I mean. Let’s say you have twenty or fifty or five hundred proteins crawling along a DNA strand. If even ONE of them stops to hump the DNA, nobody else can get by. Like camping at the bottom of an escalator. Traffic Jam! So does that mean that all transcription is “rate limited” by the slowest moving protein? Does RNA polymerase slide over the whole affair like an overhead tram? Can little proteins leapfrog, hop scotch, or cart wheel off and on the DNA? Nature is cunning, so my guess is that they not only cartwheel, but that they have fun doing it. Barry
made a wonderful remark about the proteins just sliding along “electrostatically” which
makes me want to know: If that little ligase is sitting on the track when the Queen Mary (RNA polymerase), comes along, that teensy weensy protein inhibits the synthesis of yet another. Barry drew a big circle on the paper and marked the -35 base, -10 base and + 1 base positions as the gene relative markers that the RNA transcriptase. The presence of a protein on those tracks inhibit expression. Happily this brought up the issue of length and “junk DNA”. The stretches between the -35 position and the -10 position could be filled with ANYTHING YOU WANT, as long as the LENGTH relationship were preserved. So two kinds of relationships are being encoded. Instructions and spacers. Spacers are 1 dimensional packaging. Therefore some of our DNA contains the “how to make a protein” information and some of it POSITIONS THE INFORMATION structurally (or biophysically) Is that really really true? Is that always true? Inquiring minds want to know! Could you stash secret messages in there to be used later for something else or is spacer function always distinct from instruction function? Does spacer information ever encode species attributes like, “how long my forearm is”? How is full grown organism structural geometry mapped on to the gene? How the heck did that get run length encoded into an instruction that was only 7 Gigabits long? (3.5 Gigabases in base four requires 7 Gigabits to represent) Interesting fact. E. Coli DNA is about a thousand times simpler than we are ( roughly 3.5 Megabases as opposed to 3.5 Gigabases) But if you look at a person, gross anatomy, microanatomy, they seem a lot more than a 1000 times more complex. There are a 100 million neurons in the brain alone, which itself is woven into an intricate and multileveled architecture. Support cells that makes a trillion element brain, and the whole body is a 100 trillion. Whew! Then we are talking coiling, supercoiling and BENDING. I asked Barry if transcription could be inhibited by the density of the coiling. I wonder if 23 chromosomes is a packaging issue based on sequential access. It would be easier to get around the rate limiting business if there were 23 parallel tracks, only that doesn’t answer why the 23 tracks are all different lengths, nor does it answer why there aren’t 2300, or 23000 chromosomes which would keep the traffic jams down. Packaging led to recursion and recursion to levels of detail. That there are multiple levels of coiling, and there are biophysical/biochemical implications of those multiple levels that correspond to the need both to package and to control expression. Uncoil this stuff and its about 6 feet long. To big for my suitcase!. So,
wrapping up, there are more pointed questions in categories
that look like: I
had to go to the dentist, wish I could have kept learning.
We parted talking about proteins with dual roles, how exquisite
the design was even for viruses and bacteria, all the way to
the core. I asked him if we could pick up on that theme next
time… - Van |
|