|
9/8/99
|
HP Dr. Drake
|
Biochemistry
|
Summary Notes
|
L. Van Warren
|
|
Chapter 6
Covalent
Structures of Proteins
|
|
- Proteins, such as rhodopsin in the retina, acquire
sensory information that is processed through the action of nerve cell proteins.
- The proteins of the immune system, such as the
immunoglobulins, form an essential biological defense system.
- Proteins such as collagen play structural support
roles.
- Protein function can only be understood in terms
of protein structure.
1) A protein's primary structure is the amino
acid sequence.
2) A proteins secondary structure is the local
spatial arrangement of polypeptide's backbone atoms without regards to the
conformation of its side chains.
3) Tertiary structure refers to the three-dimensional
structure of the entire polypeptide.
1.
Primary Structure Determination
- Insulin was sequenced by Sanger in 1953, 51
residues long, > 100 g required.
- b-galactosidase,
1021 residues long 1978
- Edman degradation
- phenylisothiocyanate + polypeptide ->
phenylisothiocarbamyl adduct
- phenylisothiocarbamyl adduct + trifluoroacetic
acid -> released residue
- Edman takes the first element of the list off
on the N-terminal end, and leaves the rest of the list intact. Edman is therefore
a computational protein operator. We can write this as:
List = First(List) + Rest(List)
NPolypeptide = N + Polypeptide
Edman(NPolypeptide) = N + Polypeptide
- Edman degradation can be replied repeatedly until
the Polypeptide is completely sequenced, and is thus amenable to automation.
- C-Terminus Identification
- Done Using Exopeptidases.
- Carboxypeptidase A works provided that Rn !=
Arginine, Lysine, or Proline and Rn-1 != Proline. These relationships are
summarized below:
B. Cleavage of Disulfide Bonds
- Permits separation of polypeptide chains if they
are connected via Cysteine disulfide bonds, e.g.
T D I Q G C W W H C S
. . . . . |
R Q E I A C D
- To expose internal protein structures to proteolytic
(protein-cleaving) agents for further processing.
- Cleave disulfide bonds with performic acid
- This converts S-S bridges (in Cys-Cys) form to
cysteic acid
- but messes up Met residues
- A better way is 2-mercaptoethanol, dithiothreitol
or dithioerythritol (Cleland's reagent).
- Iodoacetic acid is used to alkylate free sulfhydrl
groups to prevent renaturation of disulfide bonds by free oxygen in the air.
C. Separation, Purification and
Characterization
of the Polypeptide
Chains
- Urea and guanidinium ion are denaturing agents.
D. Amino Acid Composition
- Before sequencing we determine amino acid composition.
- Amino Acid composition is determined by complete
hydrolysis followed by quantitative analysis of the liberated amino acids.
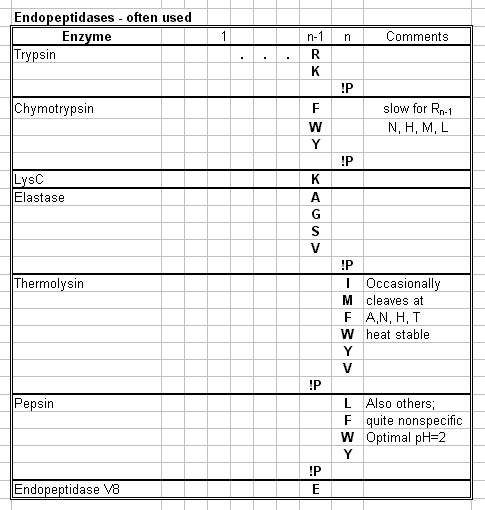
- Endopeptidases are used for hydrolysis.
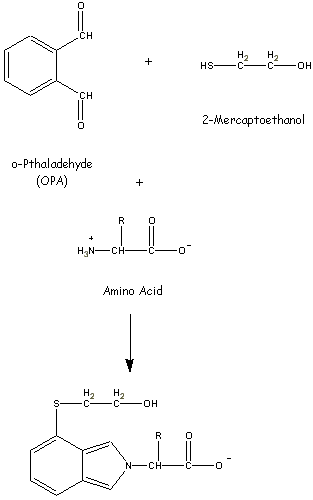
- Amino Acid Analysis Has Been Automated
- AA's are separated via ion exchange chromatography
and visualized using fluorescent adducts.
- The Amino Acid Compositions of Proteins Are Indicative
of Their Structures
- The amino acid analysis of a vast number of proteins
indicates that they have considerable variation with respect to their amino
acid compositions.
- Polar/Nonpolar > 1 for globular proteins
- Nonpolar residues predominate in membrane-bound
proteins!
E. Specific Peptide Cleavage Reactions
- Polypeptides that are longer than 40 to 80 residues
cannot be directly sequenced.
- Polypeptides of greater length must be cleaved
enzymatically or chemically into fragments small enough to be sequenced.
- Trypsin Cleaves Peptide Bonds AFTER positively
charged residues N and K if
Rn != P
- Cyanogen Bromide Specifically Cleaves Peptide
Bonds after Met Residues
F. Separation and Purification of Peptide
Fragments
- Reverse-Phase Chromatography via HPLC has reduced
separation of peptide fragments to routine procedure.
G. Sequence Determination
- Repeated Edman Degradation in a sequenator accomplishes
sequencing.
- Usually, a peptide's 40 to 60 N-terminal residues
can be identified
(100 or more with the most advanced systems)
before the cumulative effects of incomplete reactions, side reactions and
peptide loss make further amino acid identification unreliable.
H. Ordering the Peptide Fragments
- Peptide fragments are ordered by comparing amino
acid sequences of one set of peptide fragments with those of a second set
whose specific cleavage sites overlap those of the first set.
I. Assignment of Disulfide Bond Positions
- Disulfide Bond Positions can be determined by
diagonal electrophoresis.
J. Peptide Sequencing by Mass Spectrometry
- Good for up to 25 residues.
- Fast Atom Bombardment in concert with Tandem
Mass Spectrometer
- Good for mixtures of peptides
K. Peptide Mapping
- Related proteins can be sequenced more rapidly
once a primary structure has been solved.
- This is called fingerprinting or peptide mapping.
L. Nucleic Acid Sequencing
- It is easier to sequence DNA than proteins BUT:
- Disulfide bonds can be located only by protein
sequencing.
- Many proteins are modified after biosynthesis
- It is sometimes difficult to locate the nucleic
acid that encodes the protein of interest. (that will change with complete
human genome project)
- A common DNA sequencing error is the insertion
or deletion of a single CTAG.
- The standard genetic code is not universal.
2.
Protein Modification
- A common strategy for identifying proteins is
to use side chain specific reagents.
- There is a big table of these in the book.
3.
Chemical Evolution
- A mutation in a protein must increase the probability
of survival.
A. Sickle Cell Anemia: The Influence of
Natural Selection
- Hemoglobin is a protein for oxygen transport.
- a2b2
tetramer
- two identical alpha chains and two identical
beta chains.
- erythrocytes carry the hemoglobin
- Sickle-Cell Anemia is a Molecular Disease
- In 1945 Linus Pauling hypothesized that sickle-cell
anemia was the result of a mutant hemoglobin.
- Later they showed that normal hemoglobin HbA
had an anionic charge around tow units more negative than HbS.
- Glu in the sixth position of each beta chain
was changed to Valine.
- This mutation caused HbS to aggregate into filaments
of sufficient size and stiffness to deform erythrocytes.
- The Sickle-Cell Trait Confers Resistance to Malaria
- Individuals Heterozygous for HbS are resistant
to malaria.
- Sickle-Cell Anemia provides a classic Darwinian
example of a single mutation's adaptive consequences in the ongoing biological
competition among organisms for the same resources. (dogma alert)
B. Species Variations in Homologous Proteins:
The Effects of Neutral Drift
- A protein that is well adapted to its function
still continues evolving.
- Comparisons of the primary structures of homologous
proteins (evolutionarily related proteins) indicates which of the residues
are essential to function and which have lesser significance.
- Cytochrome c Is a Well-Adapted Protein.
- It is used in mitochondria as part of an electron
transport chain.
- It transfers electrons between cytochrome c reductase
and cytochrome c oxidase.
- Protein Sequence Comparisons Yield Taxonometric
Insights
- Proteins Evolve at Characteristic Rates
- The rate that mutations are accepted into a protein
depends on the extent that amino acid changes affect its function.
- Mutational Rates are Constant in Time
- Existence proof, insects versus plants versus
animals.
- Plant characteristic time frames are different
than animals or insects!
- Point mutations in DNA accumulate at a constant
rate with time via random chemical change rather than resulting from errors
in replication.
- Protein Evolution Is Not the Basis of Organismal
Evolution
- The rapid divergence of human and chimpanzee
stems from relatively few mutational changes in the segments of DNA that control
gene expression, that is how much proteins will be made, where and when.
C. Evolution through Gene Duplication
- A Gene duplication is an efficient mode of evolution
because one of the duplicated genes can evolve a new functionality through
natural selection while its counterpart continues to direct the synthesis
of presumably essential ancestral protein.
- The sequences of hemoglobin's alpha and beta
subunits and myoglobin are quite similar.
- 1) The primordial globin probably function simply
as an oxygen storage protein.
- 2) Duplication of a globin gene ~1.1 billion
years ago permitted the genes to evolve separately by a series of point mutations.
- 3) Hemoglobin's tetrameric character is a structural
feature that greatly increases its ability to transport oxygen efficiently.
- 4) In fetal mammals, oxygen is obtained from
the maternal circulation.
- 5) Human embryos in the first 8 weeks post conception
make gene-duplicated alpha and beta variant hemoglobins.
- 6) In primates, the beta chain has undergone
relatively recent duplication to form a delta chain. The alpha2delta2 hemoglobin
which is about 1% of the normal adult hemoglobin has no known unique function.
- The human genome contains relics of globin genes
that are no longer expressed.
- The three dimensional structure of a protein
and hence its function is dictated by its amino acid sequence.
4.
Polypeptide Synthesis
- The ability to manufacture polypeptides not
available in nature has enormous biochemical potential.
- 1) To investigate the properties of poly peptides
by systematically varying their side chains.
- 2) To obtain polypeptides with unique properties,
with nonstandard side chains or with isotopic labels.
- 3) To manufacture pharmacologically active polypeptides
that are biologically scarce or nonexistent. For example synthetic vaccines.
- Consider the homopolypeptides, polyglycine, polyserine
and polylysine.
- Oxytocin was the first nona-peptide (9 peptide)
synthesized.
A. Synthetic Procedures
- Polypeptides are chemically synthesized by covalently
linking amino acids one at a time.
- The amino acid being protected must have its
own alpha-amino group protected, then unprotected in a series of coupling
and deblocking steps.
- Amino Acid synthesis is easier if you anchor
the C terminus to an insoluble solid support such as beads of polystyrene
resin. This is called solid phase synthesis.
- Anchoring the chain to an inert support
- Coupling the amino acids
- Releasing the polypeptide from the resin is done
via liquid HF.
- Yield of these steps is low 0.98^2n where n is
the number of peptide bonds to be created