Abstract
|
The focus of this work is applying image processing, colorimetry and counting techniques to enhance the accuracy and prognostic tracking power of immunohistochemistry (IHC) assays of hormone receptors. As a proof of concept Her2Nu receptor assays are used in this example. Her2Nu receptor expression is increased certains kinds of breast cancer. This paper follows two prequels which defined and developing the method, Precise Digital Quantification of Retinal Angiopathy or PDQ-A, for short. This paper demonstrates a low cost method for improving the accuracy of IHC Her2Nu assays in high volume screening. The lowest cost assay of Her2Nu uses IHC while more expensive and time consuming assays use fluorescent in-situ hybridization or FISH. IHC assays are currently graded manually by pathologists, typically on a scale from 0 to 4+. This result is then frequently transmitted to clinicians as simply a "Her2Nu positive or Her2Nu negative" result. Variations in slide making conditions, staining procedures and human color perception (1 out of 8 adult males are colorblind and most pathologists are male) can cause a positive result to be reported as negative. This is clinically significant because only Her2Nu positive breast cancer patients are referred for treatment with Herceptin, a low toxicity chemotheraputic agent for metastatic breast cancer. This paper outlines a set of image processing techniques that remove subjective human estimation from the loop and replace the 0 to 4+ scale with a quantitative measurement accurate to at least two significant digits. This has two advantages. First quantitative reporting and second, disease staging and tracking. |
Resolution
Digital images have an associated spatial resolution of width by height
pixels and a colorimetric resolution of colors possible shades
given by 2^24 or approximately 16 million possible shades. These shades derive
from the combination of three channels of color, red, green and blue, in eight
bits of intensity for each channel - 3 x 8 =24. The preliminary image
in this paper was digitized at 2027 dots per inch in 24 bits color bits
in an approximate aspect ratio of 3:2 from a 35 mm slide consisting of a four
panel montage. It was then cropped according to the initial step of
the method and spatially downsampled in a 4:1 spatial ratio prior to further
analysis. The images published in this note are further decimated via
adaptive diffusion dithering method and stored for transmission using JPEG
compression. JPEG compression causes some color artifact and slight
loss of detail. This does not affect the PDQ method in any way, since
full 24 bit color resolution was maintained during image analysis.
Step One
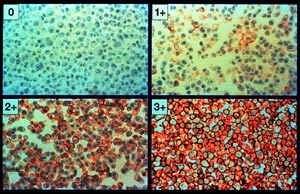
Image 1, below, shows the four panel montage:
Actual Her2nu IHC receptor assay
image
(courtesy Dr. Lee Schwartzberg, West Clinic)
In this initial experiment we are primarily interested in relative color differences rather than absolute color values. We assume that the dynamic range of the native film emultion is adequate to capture the color relationships we are attempting to characterize. A 0.46 degree CCW Rotate operator was applied prior to subsequent image processing.
Step Two
The goal of Step Two as in the prequel was to eliminate background areas
of the scan from consideration. This was done by sampling the non staining
areas and replacing values in an intensity space neighborhood with black (0,
0, 0) in red, green and blue respectively. The null color is chosen
such that it can be easily counted and subtracted from the slide area.
Black was chosen on the synthetic retina, white was used on the mouse retina
and black will be used here to record negative regions. Black is also
far in color value from regions of profound positive staining. In the
regions compared here, the darkest regions are negative, non-staining regions.
Since there are no darker regions that are not pathologic we may set this
extreme to {0, 0, 0}. The exact selection of a cutoff is greatly
facilitated by inspecting the non cell regions of the image background and
correlating the rgb values with the corresponding histograms. Unlike
the retinal counterparts in the prequel, there is not a single channel such
as green that is diagnostic since the stain ranges over a wide set of shades
that do not directly correlate with the primary image storage colors.
The key to the method is alternately eliminating background signal while enhancing Her2Nu signal.
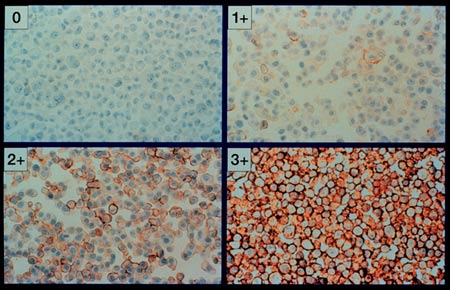
Step Three
In the next step the region of clearly positive signal is selected for enhancment.
In our case the yellowish, brownish and dark reddish areas that stained positive
are sampled and turned white. The strips to the left of each image section
retain their original coloration. This took
approximately 2 seconds on a 900 MHz processor running the Windows operating
system.
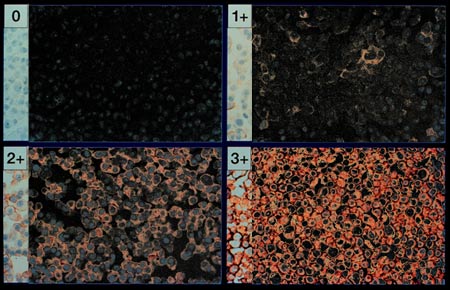
Step Four
In this step, we take the remaining cell walls that do not stain positive
and reduce their significance to zero. The remaining lighter red areas are
taken to white. Thus the negatively stained regions are completely black and
positively stained regions are shades of light gray and white.
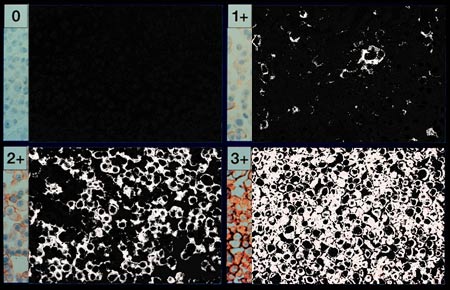
A modification of this method could enable the actual Her2Nu receptor count to be estimated. If this was applied to other hormone and receptor families one could conceivably do a "receptor inventory" of tissue specific cell lines. This might be useful in characterizing and differentiating neoplastic cell lines.
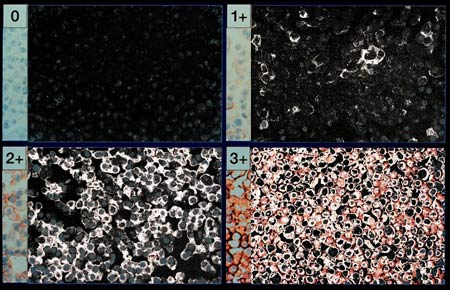
Step Five
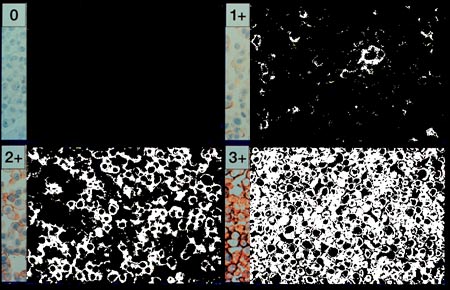
We are now at a pivotal and fascinating point in the process.
When we began, we had an image in which it was difficult for a person to
quantify the regions of Her2Nu positive status. By moving the negative
pixels to black and positive pixels to white, we now have an image which is
more readily assessed and can be exactly counted. The last image processing
step is to push the light gray and white pixels to complete whiteness. This
prepares the image for pixel counting.
Step Six
The pixels are counted by direct readout of the image histogram. In each of
the four regions 100,270 pixels were considered and those that were positive
were accumulated in the numerator. The percentages are the percentages of
the sampled field that are positive in the Her2Nu IHC staining procedure.
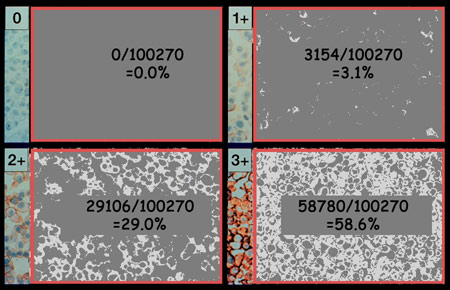
For reporting purposes and to avoid physician confusion, it would be useful to convert the percentages of image area to percentages of cell surface covered by the receptor in question.
Conclusion and Discussion
The proposed method works. The next step is to further refine the method
with a larger number of IHC assay results. As this method is applied in the
real world it would be important to include positive, negative and graduated
controls that could be used to calibrate the exact receptor count. This
could then be applied to other receptor families as a tool for cell surface
characterization, a useful technology that could be made real time and teleoperated
for CellWorld virtual
laboratories. This method has the real world benefit of increasing the precision
of the IHC assay, while keeping diagnostic costs low.
Acknowledgments
(c) 2000 L. Van Warren/Warren
Design Vision * All Rights Reserved