Cell Born Computing
Van Warren
12/99
Introduction
We don't know what the possible outcomes of this line of experimentation, but it is the process of directed searching and inquiry that produce new opportunities.
Potential uses
1) nano level cell instrumentation
cell is instrumented to disclose information as part of its housekeeping functions.
2) combinatorial problem solving
as in combinatorial chemistry, large solution spaces are searched in parallel by many cellular processors working concurrently
3) biological interface to computing systems
currently, all computer input takes place through the hands and
all computer output takes place through the eyes and ears.
biocompatible implants, such as artificial retinas are already moving in this direction, but are still somewhat clumsy and low in resolution and bandwidth
Practical examples
classic computer science problems
examine plausible biological embeddings of these problems
execute them to solution and ask
Part One
representing information in cellular systems
simple arithmetic, predicates, comparison, threads of execution
sorting and searching
Representing Information in Cellular Systems
There are two kinds of computational entity that we can represent at any level, in any system, they are the dual forms:
At the cellular level, patterns of information are represented as:
At the subcelluar level, patterns of information can be represented as:
Since we would ultimately like these methods to be human compatible and therapeutic, for our initial investigations we will set the following goals.
Non destructive write in.
Non destructive execution.
Non destructive read out.
Initial Choices
We would like to represent information the way the cells does, as DNA sequences.
We will take for our first problem the problem of representing, sorting and searching a list of names.
BNF for Ideal LALR Grammar:
Issueslist: nameUnit OR
nameUnit list
nameUnit: intron name intron
name: sixtyFourBases
intron: weDecideBasedOnNonDestructiveDoctrine
concurrent versus concatenated representations
concatenate, solve, concatenate
the reading problem
results as sequences
results as proteins
results as macro level objects
References:
http://www.hks.net/~cactus/doc/science/molecule_comp.html
http://www.cs.sandia.gov/jcb/v6/n1/v6n1art4.html
Experiment 1: Certain Preliminaries
The difficult part of the sorting and searching problem described above is designing a sorting algorithm that works in vivo. The major histocompatibility complex (MHC) performs exon shuffling, but shuffling is the opposite of sorting. We desire to create the minimal sorting operations of selection and successive positional exchange. One possible mechanism for accomplishing this is RNA intron self-splicing. Prior to embarking on this ambitious goal, it is useful to examine equilibrium structures of our name list. This was performed in the first of a series of numerical experiments outlined below.
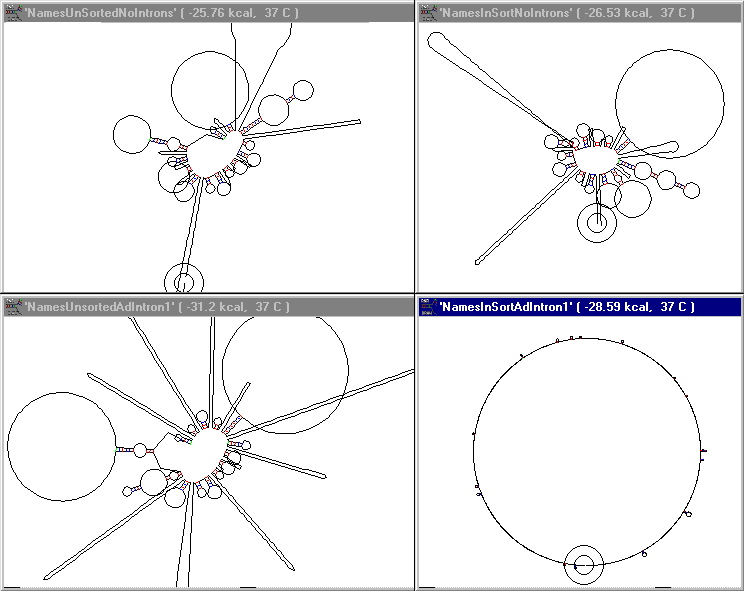
The list of 10 person names, each consisting of 16 ASCII characters was translated into an arbitrary 64 nucleotide mapping. These names are taken directly from the name representation section computed above. These are shown in their RNA secondary structures as computed by RNADraw1.06 below. The results are shown in the 2 x 2 array of images below. Proceeding from left to right shows the effect of sorting only. Proceeding from top to bottom shows the effect of adding an intron only. The intron in this naive case is a 64 nucleotide intron of the form GU-AAA...-AG. No pyrimidine rich section preceded the AG. The result is that adding introns to the unsorted list opened up but failed to geometrically delimit the final structure. Similarly presorting the list suffered from the same limitation. When presorting and adding introns occur simultaneously, the result is the significant change in secondary structure shown in the lower right frame. (widen your browser to see this...)

The titles on the images indicate conditions.
Zooming
in on the same content we can see more clearly the fine structure of the RNA
self-annealing.
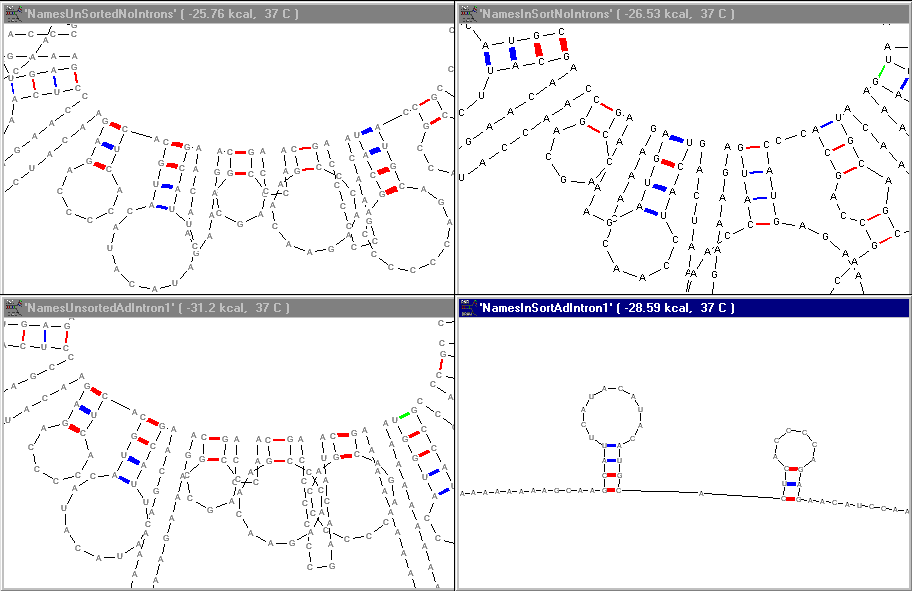
The next step will be to design introns which delineate the names in a more accessible fashion for selection and exchange.
The critical sections of a sort are:
Before we figure out how to encode these three primitive operations actions, lets improve our understanding of RNA self-annealing.
We examine the annealing patterns of 9 randomly generated RNA's, each consisting of 64 bases. Each of these patterns is unique and forms a pictograph containing 64 * 4 = 256 bits of information. There 3.4 * 10^38 of these symbols. A very large vocabulary. Six bases would give the ability to represent 1024 possible symbols. We might want to restrict certain combinations of symbols as forbidden if they had undesireable preexisting meanings to biological systems. We might want to represent symbols using multiples of three bases, since triplets form codons in genes. We might want to represent symbols using combinations of the three stop codes UAA, UAG, UGA, so that we could guarantee that no proteins would be generated.
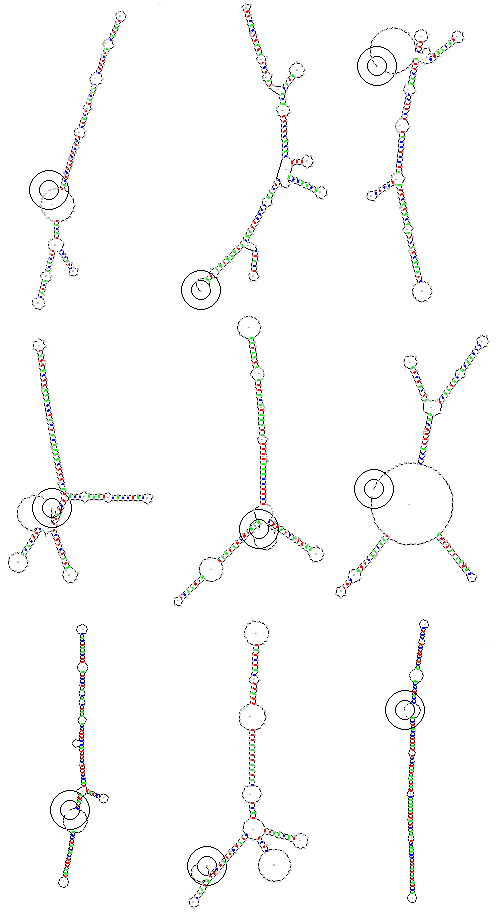
- lvw99
We might choose to have the cellular machinery serve us at one point, and then convert to a representation that would not interfere with a follow-on computation. This could be done with permutations of stop codes, which would be transcribed by RNA polymerase, but not translated by ribosomes creating proteins that would risk damaging the cell. This is consistent with our non damaging doctrine stated above.
By using only stop codes, we have lost some of the economy of a terse representation of information, but we have greatly gained in our ability to express safe computation. We are now ready to design a refined grammar. With six places in a base 3 system (from the 3 stop codes representing 0, 1, and 2 respectively) we can represent 243 characters, which is the entire ASCII character set plus change. This will require six places consisting of 18 actual bases. To represent a 16 letter name will require 288 bases. Attempts to shorten this are unproductive since five places can only represent 81 characters but still requires 240 bases to represent a 16 letter name. A simulation of these choices is attached, use the Excel sheet labeled Combinations. In the next section this method will be applied to the name list and the alphabet of RNA's will be produced.